SBML models for KEGG
All SBML files in this directory were converted from KEGG (Kyoto Encyclopedia on Genes and Genomes) by using our conversion script kegg2sbml with the permission of Professor Minoru Kanehisa (Kyoto University).
The conversion was performed by Akiya Jouraku, Nobuyuki Ohta, Akira Funahashi and Hiroaki Kitano, as part of the JST ERATO-SORST Kitano Symbiotic Systems Project. Publication of papers and other documents based on this database must include proper acknowledgment of this fact.
SBML models for KEGG Metabolic Pathways (Released 2008.08.09)
Source Databases:
* KEGG Metabolic Pathways Release 47
*
LIGAND 2008-07-28 (daily snapshot)
Downloadable tarballs (72,095 SBML files cotained in each tarball):
* SBML files without CellDesigner's tag: [Level-2 Ver-1 | Level-2
Ver-3]
* SBML files with CellDesigner's tag:
[Level-2 Ver-1]
[previous tarballs dated 2004-6-16]
https://www.genome.jp/kegg/index.html
http://sbml.org/Software/KEGG2SBML.html
http://www.symbio.jst.go.jp/symbio2.html/
posted 2008.08.07
BioModels Database

[Le Novre N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H,
Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M. BioModels
Database: A Free, Centralized Database of Curated, Published, Quantitative
Kinetic Models of Biochemical and Cellular Systems. Nucleic Acids Res., in
the press]
http://www.ebi.ac.uk/biomodels/
http://www.biomodels.net/
posted 2008.03.27
CellML repository

posted 2008.03.27
PANTHER Pathways
PANTHER Pathways consists of over 165, primarily signaling, pathways, each
with subfamilies and protein sequences mapped to individual pathway
components. A component is usually a single protein in a given organism, but
multiple proteins can sometimes play the same role. Pathways are drawn using
CellDesigner software, capturing molecular level events in both signaling
and metabolic pathways, and can be exported in SBML format. Pathway diagrams
are interactive and include tools for visualizing gene expression data in
the context of the diagrams.
http://www.pantherdb.org/pathway/
posted 2008.03.27
Reactome
The Reactome project is a collaboration among Cold Spring Harbor Laboratory, The European Bioinformatics Institute, and The Gene Ontology Consortium to develop a curated resource of core pathways and reactions in human biology. The information in this database is authored by biological researchers with expertise in their fields, maintained by the Reactome editorial staff, and cross-referenced with the sequence databases at NCBI, Ensembl and UniProt, the UCSC Genome Browser , HapMap, KEGG (Gene and Compound), ChEBI, PubMed and GO. In addition to curated human events, inferred orthologous events in 22 non-human species including mouse, rat, chicken, puffer fish, worm, fly, yeast, two plants and E.coli are also available. A description of Reactome has been published in Genome Biology.
https://reactome.org/
posted 2008.03.27
JWS Online
JWS Online is One of the first resources offering curation of the models it distributes, and online simulation. It is linked to journals such as Microbiology, FEBS Journal and IEE Proceedings in Systems Biology, that deposit the models upon submission of the manuscripts, so as to make them available to the reviewers. It now distributes the models in SBML and Pysces formats.
[Olivier BG, Snoep JL. Web-based kinetic modelling using JWS Online.(2004) Bioinformatics, 20: 2143-2144]
https://jjj.biochem.sun.ac.za/
posted 2008.03.27
The Database of Quantitative Cellular Signalling (DOQCS)
The Database of Quantitative Cellular Signalling (DOQCS) is a repository of models of signalling pathways present in the neurons. It includes reaction schemes, concentrations, rate constants, as well as annotations on the models. The database provides a range of search, navigation, and comparison functions. The pathways can be downloaded in the format used by the neuronal simulator GENESIS.
[Sivakumaran S, Hariharaputran S, Mishra J, Bhalla US. The Database of Quantitative Cellular Signaling: management and analysis of chemical kinetic models of signaling networks. (2003) Bioinformatics, 19: 408-415]
https://doqcs.ncbs.res.in/
posted 2008.03.27
SigPath
SigPath is an interesting project to develop an open knowledgebase of qualitative pathways and quantitative models related to signalling. An interesting feature is the possibility of annotating model components.
[Campagne F, Neves S, Chang CW, Skrabanek L, Ram PT, Iyengar R, Weinstein H. Quantitative information management for the biochemical computation of cellular networks. (2004) Science STKE, 248: PL11]
http://www.sigpath.org/
posted 2008.03.27
ModelDB
ModelDB is being developed as part of the SenseLab effort, the resource distributes models encoded in the many different formats, mainly those used by the GENESIS and NEURON simulators, but also format used by generic simulation environments such as Octave, MatLab, Octave or XPP-Aut.
https://senselab.med.yale.edu/modeldb/
posted 2008.03.27
PathArt
PathArt is a comprehensive collection of manually curated information from literature as well as public domain databases on more than 1000 signaling and metabolic pathways.
https://www.jubilantbiosys.com/pd.htm
posted 2008.03.27
CellDesigner models
Here are the models created by CellDesigner.
SBI Model Base
A series of molecular interaction maps created by SBI using CellDesigner.
Metabolic Syndrome
Molecular Interaction Map of Macrophage
EGFR Pathway Map
Yeast Map
Graphical Notation Samples
posted 2008.03.27
SIGMOID
The SIGMOID project is intended to produce a database of cellular signaling pathways and models thereof, to marshall the major forms of data and knowledge required as input to cellular modeling software and also to organize the outputs. Such cellular signaling and regulatory pathways are commonly hand-drawn in biological literature as an aid to intuitive understanding. Pathway databases can provide the same assistance in the context of attempts to achieve a quantitative understanding of cellular processes by numerical simulation. They can also serve as an aid to capturing and querying both expert knowledge and heterogeneous data sets pertaining to pathways. Cell model databases are a subject of current research. SIGMOID works at the interface of these two areas.
http://sigmoid.sf.net/
posted 2007.08.01
Toll-Like Receptor signaling network
The model and the description is available at Molecular Systems Biology
Oda,K.; Kitano,H.;
A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol : msb4100057. Apr. 18, 2006

Operating Environment:
CellDesigner 2.2
Data links to www.nature.com/msb site:
- Map in PDF format
- Map in XML format (created by CellDesigner 2.2)
*SBML with CellDesigner annotations. PubMed IDs are embedded in the notes of the reactions/components.
posted 2006.04.18
Graphical Notation Samples
SBML files for some of figures appeared in Kitano, et al., Nature Biotechnology, August 2005.

Sample Files used in Figures:
- CellDesigner 3.0 alpha version
*new graphical notation preview version. - CellDesigner 2.2 version
posted 2005.08.01
EGFR Pathway Map
Epidermal Growth Factor Receptor Pathway Map Ver.2 (2005/5/24) The model and the description is available at Molecular Systems Biology
Oda,K.; Matsuoka,Y.;Funahashi,A.;Kitano,H.;
A comprehensive pathway map of epidermal growth factor receptor signaling. May, 2005.
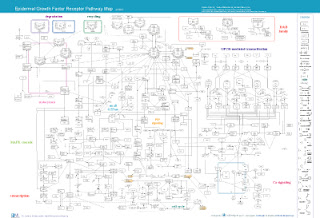
Operating Environment:
CellDesigner 2.2
Data links to www.nature.com/msb site:
- Map in PDF format
- Map in XML format (created by CellDesigner 2.2)
*SBML with CellDesigner annotations. PubMed IDs are embedded in the notes of the reactions/components.
Epidermal Growth Factor Receptor Pathway Map Ver.1 (2005/1/17)
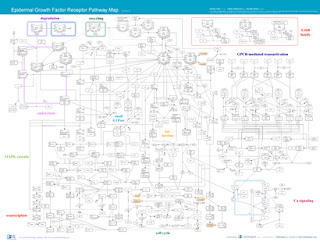
- Map in PDF format
- Map in XML format (created by CellDesigner 2.0)
*SBML with CellDesigner annotations
posted 2005.05.24
Macrophage Map
Molecular Interaction Map of Macrophage
A team of researchers at the Systems Biology Institute (SBI), Tokyo Medical and Dental University (TMDU), and Keio University jointly created a large scale molecular interaction map of macrophage. The map is intended to serve analysis framework for the Alliance for Cellular Signaling (AfCS), a large scale NIH-funded project lead by Professor Alfred Gilman (Univ. Texas Southwestern Medical Center). The map and description of the map is available from
Nature Signaling Gateway.
http://www.signaling-gateway.org/reports/v2/DA0014/MacrophageMolecularInteractionMapVer1.0.pdf

REPORT:
Molecular Interaction Map of a Macrophage
Oda K, Kimura T, Matsuoka Y, Funahashi A, Muramatsu M, Kitano H
August 25, 2004
Research Report
new Release Name: Molecular
Interaction Map of Macrophage Ver.2.0 (released Dec 2004)
Features:
Toll-like receptors (TLRs) signalings via myeloid differentiation primary response gene 88 (MyD88) -dependent and MyD88-independent pathways are added. As input signals, we selected triacyl lipopeptide for TLR1/TLR2 complex, double-stranded RNA (ddRNA) for TLR3, flagellin for TLR5, lipoprotein for TLR2/TLR6 complex, single-stranded RNA (ssRNA) for TLR7, nonmethylated CpG DNA for TLR9. (Reference: Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. J Leukoc Biol. 2004 Sep;76(3):514-9.)
Operating Environment:
CellDesigner version 2.0
Release Name: Molecular Interaction Map of Macrophage Ver.1.0
(released: Jun 2004)
Features:
This map was created with CellDesigner version 2.0.
A total number of 506 reactions and 678 species were included.
** MAP in PDF format available as part of AfCS Report
Operating Environment:
CellDesigner version 2.0
posted 2004.08.25
Metabolic Syndrome
A molecular interaction map that describes most interactions involved in the metabolic syndrome for adipocyte, hepatocyte, skeletal muscle cell, and pancreatic beta cell.
The map is prepared for the following paper:
Kitano, H., Oda, K., Kimura, T., Matsuoka, Y., Csete, M., Doyle, J., Muramatsu, M..
Metabolic Syndrome and Robustness Tradeoffs, Diabetes 53, S6-S15, 2004
>Full text [PDF]
Release Name: Maps 2004A
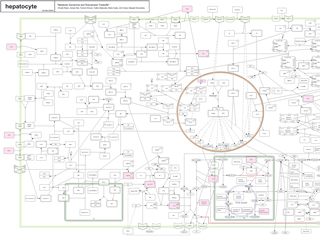
Features:
Maps are prepared in two kinds of file formats:
1) in PDF format (zipped)
2) in SBML format created by CellDesigner 2.0 (zipped)
- adipocyte
- hepatocyte
- skeletal muscle cell
- pancreatic beta cell
Operating Environment:
Please open the SBML files with
CellDesigner 2.0
PDF format>>
SBML format>>
posted 2004.08.25
 Systems Biology Portal
Systems Biology Portal